(UroToday.com) The 2025 American Society of Clinical Oncology (ASCO) Annual Meeting held in Chicago, IL between May 30th and June 3rd, 2025, was host to a prostate, testicular, and penile cancers poster session. Dr. Jacob Berchuck presented a plasma epigenomic profiling study to reveal molecular correlates of response and resistance to 177Lu-PSMA-617 in metastatic castration-resistant prostate cancer (mCRPC).
Metastatic castration-resistant prostate cancer (mCRPC) is an advanced stage of prostate cancer with poor outcomes and limited therapeutic options. The FDA-approved radiopharmaceutical therapy 177Lu-PSMA-617 targets PSMA, offering a novel approach for mCRPC treatment. However, response to therapy is heterogeneous, and resistance mechanisms remain poorly understood. Benchmarking molecular signatures to clinical outcomes could provide critical insights into predicting response and resistance to therapy.
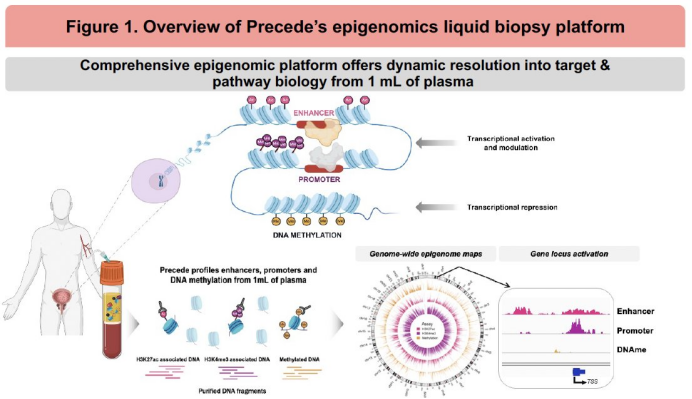
The study investigators applied a multimodal epigenomic liquid biopsy platform to profile tumor-specific transcriptional activation and resistance pathways in plasma from patients treated with 177Lu-PSMA-617.
Baseline plasma samples were collected from patients with mCRPC at the time of PSMA PET imaging and initiation of 177Lu-PSMA-617 therapy. Epigenomic profiling of genome-wide signals from promoters, enhancers, and DNA methylation was performed on 1mL of plasma (N=81, ctDNA ≥0.5%).
The study investigators tested the association of androgen receptor (AR) activity, neuroendocrine-ness (NEPC-ness), and pathway activities with response using Cox proportional hazards models. Androgen receptor (AR) activity was assessed by enhancer signals at AR binding sites, NEPC-ness was assessed via the gene-activity of NEPC marker genes, and pathway activities were calculated using gene set variation analysis (GSVA) scores based on gene proximal epigenomic signals. Response to 177Lu-PSMA- 617 was assessed by clinical radiographic progression-free survival (crPFS), PSA-PFS, time to next treatment (TTNT), and overall survival (OS). Epigenomic profiling was restricted to plasma samples with ctDNA ≥0.5%, with enhanced statistical modeling for samples with ctDNA ≥3% (n = subset used for gene-level prediction).
A total of 81 patients with PSMA PET–positive mCRPC initiating 177Lu-PSMA-617 therapy were included. Baseline clinical characteristics were as follows:
Median OS: 260 days
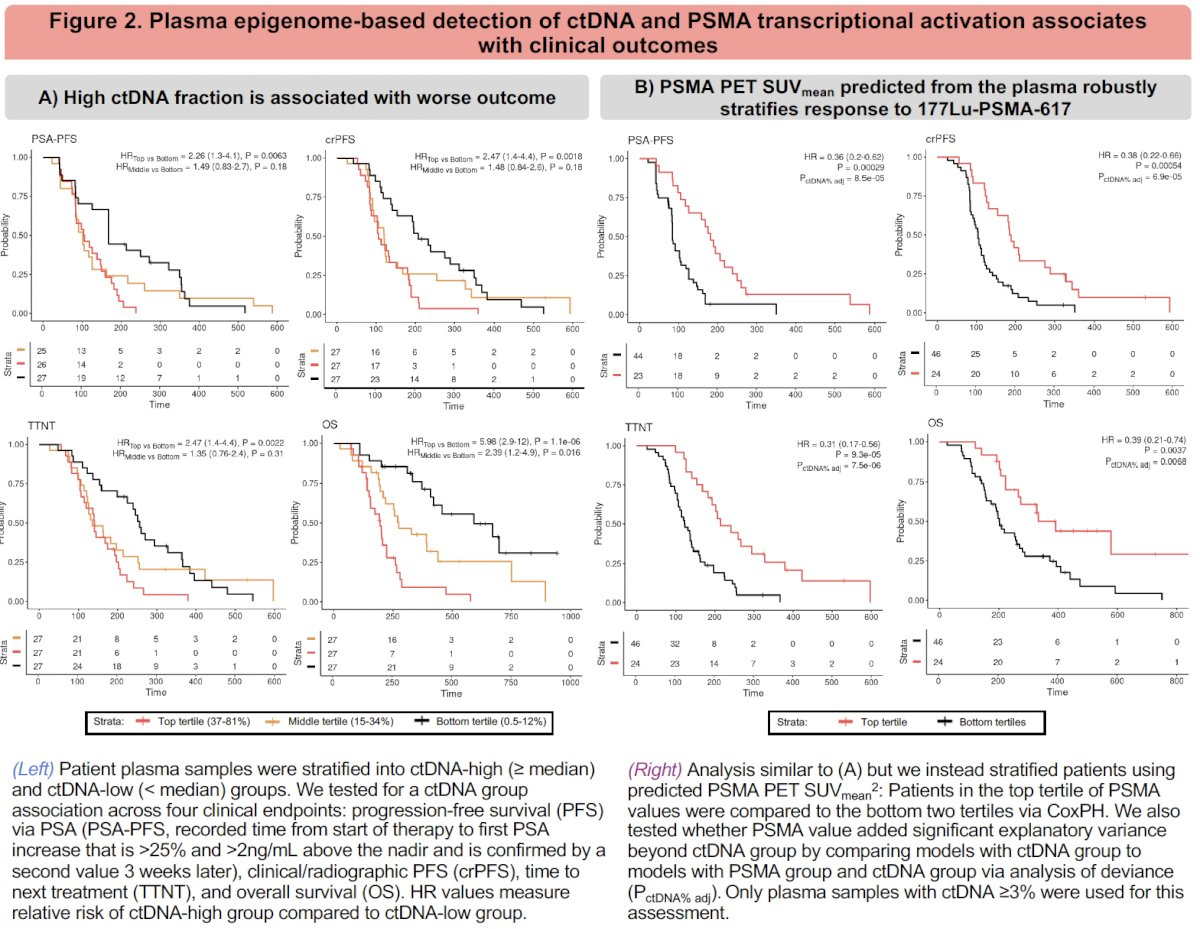
1. ctDNA Fraction Stratifies Prognosis
Patients with high ctDNA burden (≥ median) had significantly worse outcomes across all clinical endpoints:
These findings underscore ctDNA as a robust prognostic biomarker.
2. Plasma-derived PSMA Expression Adds Independent Predictive Value
The investigators trained models to predict PSMA (FOLH1) transcriptional activity from cfDNA enhancer/promoter signal. PSMA PET SUVmean predicted from plasma epigenomics robustly stratified patients for:
These findings suggest that transcriptionally active PSMA, as inferred from plasma, captures biologically relevant treatment susceptibility.
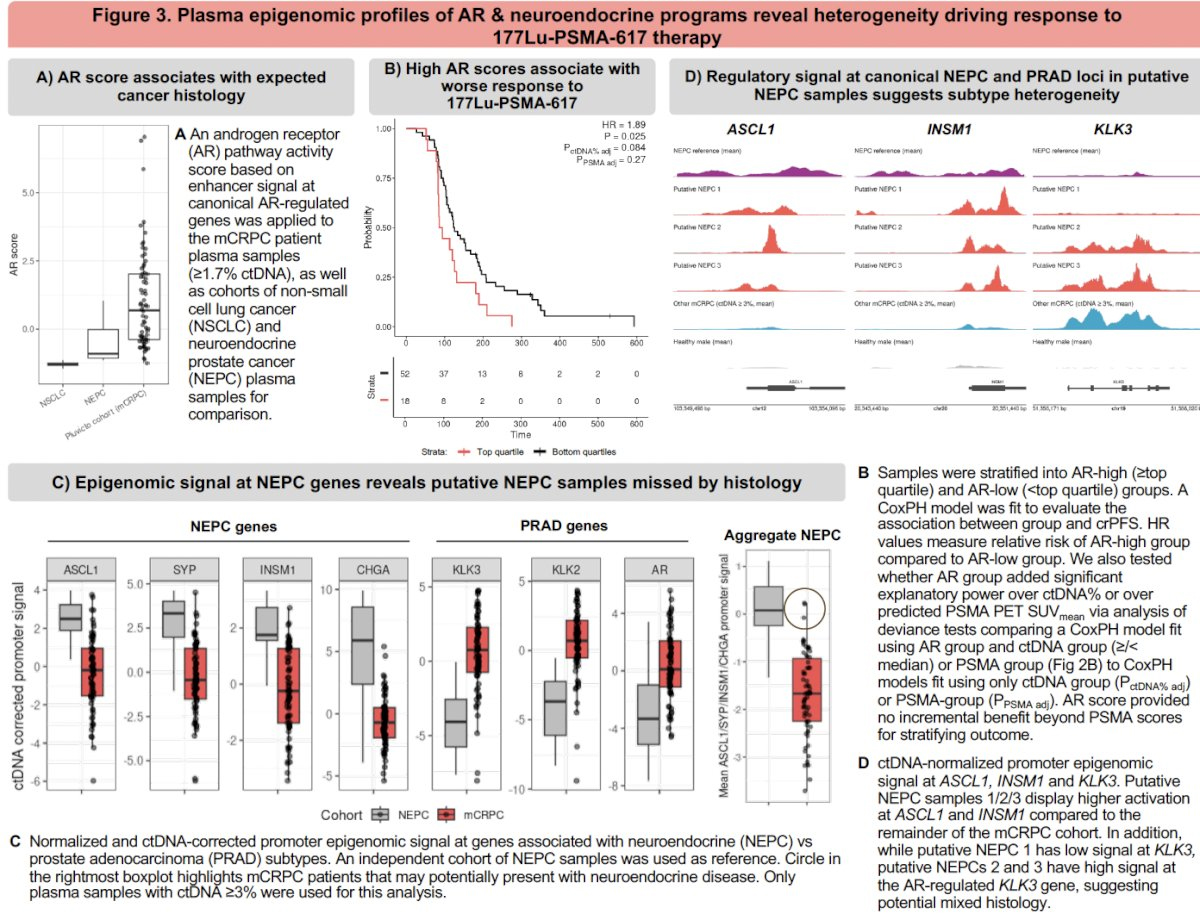
3. AR Signaling: Prognostic but Redundant with PSMA
An AR activity score based on enhancer activation at canonical AR-responsive genes was calculated. AR-high tumors (≥ top quartile) had:
However, AR score did not add independent predictive value over PSMA signal:
This suggests that while AR signaling aligns with canonical prostate adenocarcinoma biology, it may not offer incremental prognostic utility in this treatment context.
4. Identification of Neuroendocrine Phenotypes Missed by Histology
NEPC-ness was assessed using enhancer activity at ASCL1, INSM1 (NEPC genes), and KLK3 (AR-regulated). A subset of patients exhibited:
These transcriptional profiles identified putative NEPC cases that were not apparent on clinical histology—underscoring the sensitivity of epigenomic readouts for detecting phenotypic plasticity and treatment resistance.
5. Transcriptional Pathway Activity Correlates with Response and Resistance
Using gene set variation analysis (GSVA), the authors evaluated pathway activity scores (MSigDb REACTOME pathways) and associated them with clinical endpoints:
Pathway activities stratified patients across crPFS and TTNT endpoints and remained significant after adjusting for ctDNA fraction.

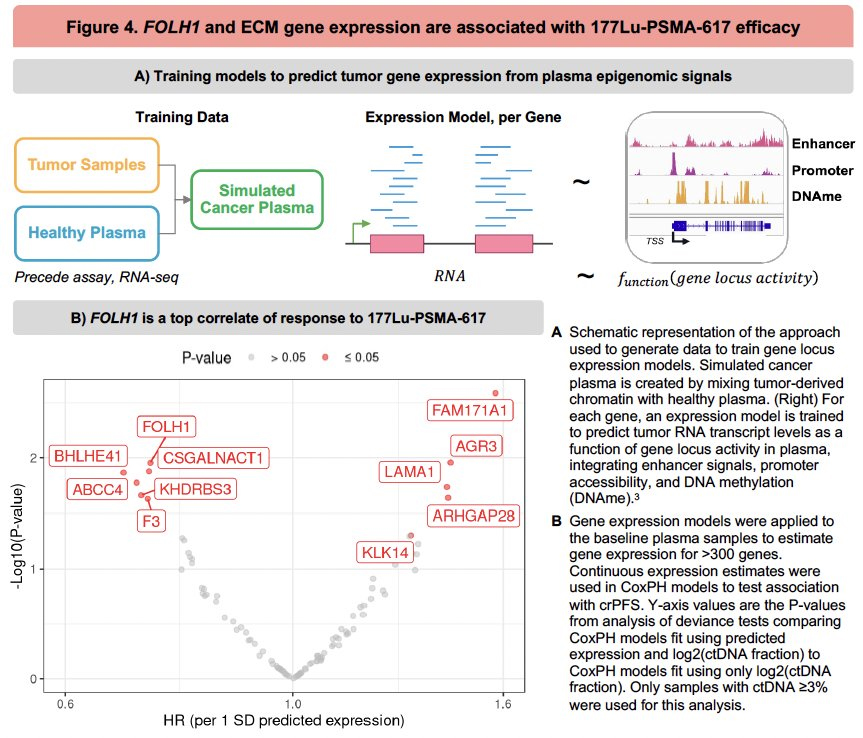
6. Gene-Level Predictive Models from cfDNA
Expression models for >300 genes were derived using synthetic cancer plasma spiked with chromatin, then applied to baseline patient samples. Notably:
Dr. Berchuk concluded as follows:
Presented by: Jacob E Berchuck, MD, Assistant Professor, Department of Hematology and Medical Oncology, Winship Cancer Institute of Emory University, Atlanta, GA
Written by: Rashid K. Sayyid, MD, MSc – Robotic Urologic Oncology Fellow at The University of Southern California, @rksayyid on Twitter during the American Society of Clinical Oncology (ASCO) 2025 Annual Meeting, Chicago, IL, Fri, May 30 – Tues, Jun 3, 2025.